Project: Predicting Titanic survival
In this project, we will be working with the very famous Titanic dataset from Kaggle.
Highlights:
- Exploratory data analysis
- SimpleImputer to impute null values
- OneHotEncoder to encode categorical variables
- Defining pipelines
- Cross validation scores
- LogisticRegression model
- RandomForestClassifier model
- XGBClassifier model
- XGBRFClassifier model
#Imports
import numpy as np
import pandas as pd
#Visualizations
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
sns.set()
#Data loading
titanic = pd.read_csv('train.csv')
#Glimpse of data
titanic.head(2)
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | Braund, Mr. Owen Harris | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S |
| 1 | 2 | 1 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C |
#drop the rows where target has missing values
titanic.dropna(axis=0,subset=['Survived'],inplace=True)
#Data information
titanic.info()
<class 'pandas.core.frame.DataFrame'>
Int64Index: 891 entries, 0 to 890
Data columns (total 12 columns):
PassengerId 891 non-null int64
Survived 891 non-null int64
Pclass 891 non-null int64
Name 891 non-null object
Sex 891 non-null object
Age 714 non-null float64
SibSp 891 non-null int64
Parch 891 non-null int64
Ticket 891 non-null object
Fare 891 non-null float64
Cabin 204 non-null object
Embarked 889 non-null object
dtypes: float64(2), int64(5), object(5)
memory usage: 90.5+ KB
We have both numerical and non-numerical data to deal with. We would have to convert the categorical features to numerical ones before we could fit our model.
#Describe numerical features
titanic.describe()
| PassengerId | Survived | Pclass | Age | SibSp | Parch | Fare | |
|---|---|---|---|---|---|---|---|
| count | 891.000000 | 891.000000 | 891.000000 | 714.000000 | 891.000000 | 891.000000 | 891.000000 |
| mean | 446.000000 | 0.383838 | 2.308642 | 29.699118 | 0.523008 | 0.381594 | 32.204208 |
| std | 257.353842 | 0.486592 | 0.836071 | 14.526497 | 1.102743 | 0.806057 | 49.693429 |
| min | 1.000000 | 0.000000 | 1.000000 | 0.420000 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 223.500000 | 0.000000 | 2.000000 | 20.125000 | 0.000000 | 0.000000 | 7.910400 |
| 50% | 446.000000 | 0.000000 | 3.000000 | 28.000000 | 0.000000 | 0.000000 | 14.454200 |
| 75% | 668.500000 | 1.000000 | 3.000000 | 38.000000 | 1.000000 | 0.000000 | 31.000000 |
| max | 891.000000 | 1.000000 | 3.000000 | 80.000000 | 8.000000 | 6.000000 | 512.329200 |
Observations:
- PassengerId holds not significant meaning and will be dropped.
- Pclass is an ordinal variable with most of the people in 3rd class
- Age seems to have null values, we will have to fill them in with some value
- SibSp is also an ordinal variable with most of the people travelling without siblings or spouses
- Parch is also an ordinal variable with most of the people travelling without their parents or children
- Max fare seems an outlier
#Describe categorical features
titanic.describe(include=['O'])
| Name | Sex | Ticket | Cabin | Embarked | |
|---|---|---|---|---|---|
| count | 891 | 891 | 891 | 204 | 889 |
| unique | 891 | 2 | 681 | 147 | 3 |
| top | Thomas, Master. Assad Alexander | male | 347082 | C23 C25 C27 | S |
| freq | 1 | 577 | 7 | 4 | 644 |
Observations:
- We can assume that name holds no significance in the survival of a passenger, hence we will drop this column
- Sex will be included in our predictions
- Looks like some people were sharing tickets. We will drop this column as well as it doesn’t show any significance
- Cabin seems to have alot of null values. We will drop it as well
- Embarked will be used in predictions
Exploratory data analysis
#Lets check for null values
plt.figure(figsize=(10,5))
sns.heatmap(titanic.isnull(),cbar=False,yticklabels=False,cmap='viridis')
<matplotlib.axes._subplots.AxesSubplot at 0x28dd1756940>
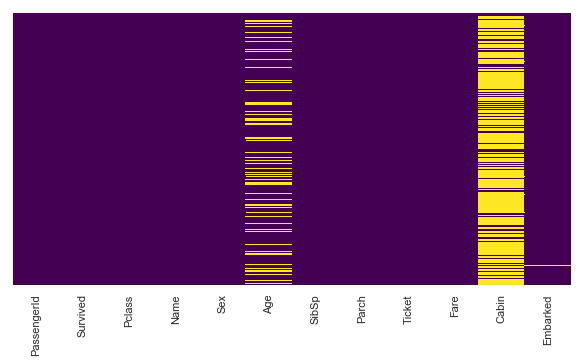
[col for col in titanic.columns if titanic[col].isnull().any()]
['Age', 'Cabin', 'Embarked']
#We will drop cabin column as it has alot of null values. Dropping all unnecessary columns
titanic.drop(['Cabin','PassengerId','Name','Ticket'],axis=1,inplace=True)
We will impute the Age and Embarked columns later.
sns.countplot(x=titanic['Survived'],data=titanic,hue='Sex')
<matplotlib.axes._subplots.AxesSubplot at 0x28dd1ab0978>
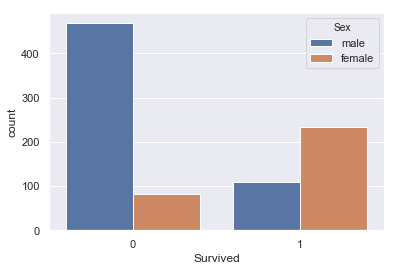
Looks like there were more female survivors.
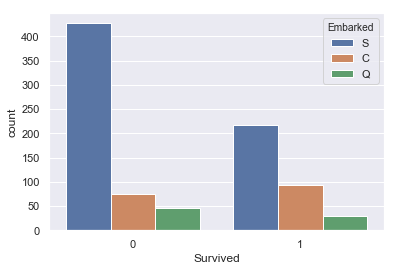
sns.countplot(x=titanic['Survived'],data=titanic,hue='Embarked')
<matplotlib.axes._subplots.AxesSubplot at 0x28dd1b03278>
sns.countplot(x=titanic['Survived'],data=titanic,hue='Pclass')
<matplotlib.axes._subplots.AxesSubplot at 0x28dd1b69470>
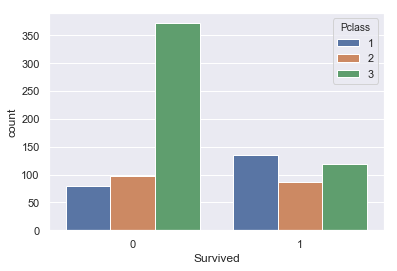
Looks like, people from 3rd class died more. Also, survival rate of 1st class is, somewhat, more than the other classes.
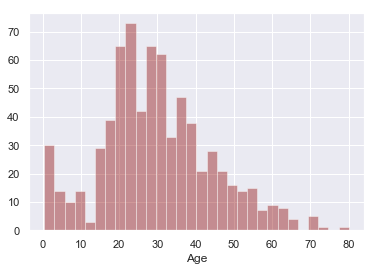
sns.distplot(titanic['Age'].dropna(),kde=False,color='darkred',bins=30)
<matplotlib.axes._subplots.AxesSubplot at 0x28dd1bd1c18>
g = sns.FacetGrid(titanic,col='Survived',row='Sex')
g.map(plt.hist,'Age')
<seaborn.axisgrid.FacetGrid at 0x28dd1add160>
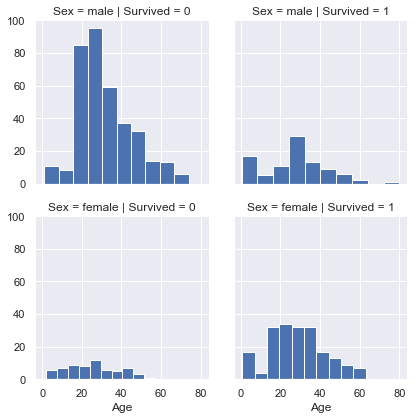
More infants survived. Also, 80 years old guy survived.
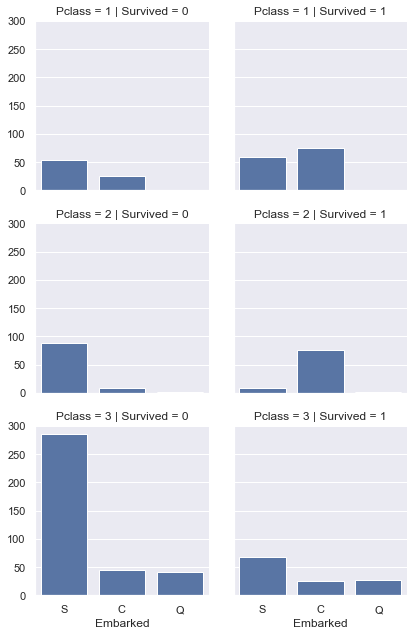
g = sns.FacetGrid(titanic,col='Survived',row='Pclass')
g.map(sns.countplot,'Embarked')
<seaborn.axisgrid.FacetGrid at 0x28dd1c6df60>
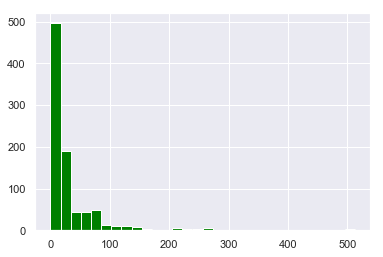
titanic['Fare'].hist(bins=30,color='green')
<matplotlib.axes._subplots.AxesSubplot at 0x28dd2155c50>
Pipelines
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import OneHotEncoder
from sklearn.pipeline import Pipeline
from sklearn.compose import ColumnTransformer
#Separate target from predictors
X = titanic.drop('Survived',axis=1)
y = titanic['Survived']
#Lets separate the numerical and categorical columns
num_cols = [col for col in X.columns if titanic[col].dtype in ['int64','float64']]
cat_cols = [col for col in X.columns if titanic[col].dtype == 'object']
#Numerical transformer
num_transformer = SimpleImputer(strategy='mean')
#Categorical transformer
cat_transformer = Pipeline(steps=[
('impute',SimpleImputer(strategy='most_frequent')),
('onehot',OneHotEncoder(handle_unknown='ignore',sparse=False))
])
preprocessor = ColumnTransformer(transformers=[
('num',num_transformer,num_cols),
('cat',cat_transformer,cat_cols)
])
Get Model
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
model1 = LogisticRegression(solver='liblinear',random_state=1)
model2 = RandomForestClassifier(n_estimators=250,random_state=1)
#Model1 pipeline
final_pipeline_1 = Pipeline(steps=[
('preprocessor',preprocessor),
('model',model1)
])
#Model2 pipeline
final_pipeline_2 = Pipeline(steps=[
('preprocessor',preprocessor),
('model',model2)
])
Cross-Validation scores
from sklearn.model_selection import cross_val_score
scores_1 = cross_val_score(final_pipeline_1,X,y,cv=5,scoring='accuracy')
scores_1.mean()
0.7912852282814269
scores_2 = cross_val_score(final_pipeline_2,X,y,cv=5,scoring='accuracy')
scores_2.mean()
0.8115290623015141
We have achieved an accuracy of around 81% using RandomForest model.
XGBoost
from xgboost import XGBClassifier,XGBRFClassifier
from sklearn.model_selection import train_test_split
X_train, X_valid, y_train, y_valid = train_test_split(X, y, test_size=0.2, random_state=1)
model_XGBClassifier = XGBClassifier(n_estimators=1000, learning_rate=0.05, n_jobs=4)
model_XGBRClassifier = XGBRFClassifier(n_estimators=1000, learning_rate=0.05, n_jobs=4)
#Pipeline #1
pipeline_XGB = Pipeline(steps=[
('preprocessor',preprocessor),
('model',model_XGBClassifier)
])
#Pipeline #2
pipeline_XGBRF = Pipeline(steps=[
('preprocessor',preprocessor),
('model',model_XGBRClassifier)
])
params = {'model__early_stopping_rounds':5,
'model__verbose':False,
'model__eval_set':[(X_valid,y_valid)]}
scores_XGB = cross_val_score(pipeline_XGB,X_train,y_train,cv=5,scoring='accuracy')
scores_XGB.mean()
0.8020450232017508
scores_XGBRF = cross_val_score(pipeline_XGBRF,X_train,y_train,cv=5,scoring='accuracy')
scores_XGBRF.mean()
0.8356706923083909
Using the RandomForest flavor of XGBoost, we were able to achieve the accuracy of around 83.5%.
Leave a comment